Outline of the Research
Natural products synthesized by microorganisms and plants in nature are very important research targets not only for elucidation of their molecular targets and signal transduction mechanisms in living organisms, but also for application to drug discovery. Since Fleming's discovery of penicillin, many natural products have been discovered in Japan from microorganisms living in the soil, and the medicines such as the antibiotic kanamycin, the anti-parasitic drug ivermectin, and the immunosuppressant FK506 have been developed based on these compounds, bringing innovation to medicine. These natural products have complex chemical structures, which are elaborated from simple precursors common to almost all living organisms by a process in which a number of enzymes (called biosynthetic enzymes) work in concert to catalyze an elaborate series of multi-step reactions (Figure 1).
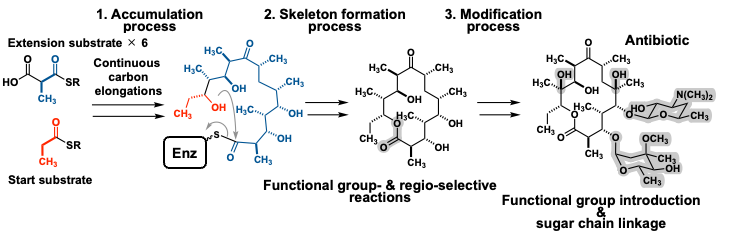
Recent advances in genome science have made available a large amount of genetic information related to the biosynthesis of natural products. On the other hand, since it has been difficult to analyze and predict the structure, reactivity, and selectivity of biosynthetic enzymes, which are gene products, many biosynthetic pathways of natural products on the earth remain unknown or untouched at present. Therefore, significant changes are needed to make effective use of these unutilized resources. Therefore, we have decided to establish this Research Area, "Forecasting Biosynthesis”, to promote organic and complementary collaborative research in close collaboration with each other based on the three main research areas of “Accumulation" (A01), “Forecasting" (A02), and “Creation" (A03) of biological reactions related to natural products (Figure 2). By freely integrating the two experimental disciplines of synthetic biology and synthetic organic chemistry, and by working closely with computational science in the information and physical chemistry fields, we can fully utilize genome information, the blueprint of natural products, as a valuable natural resource, and open up an innovative field of "biosynthetic science" that can create molecules at will. Therefore, we decided to conduct this study.
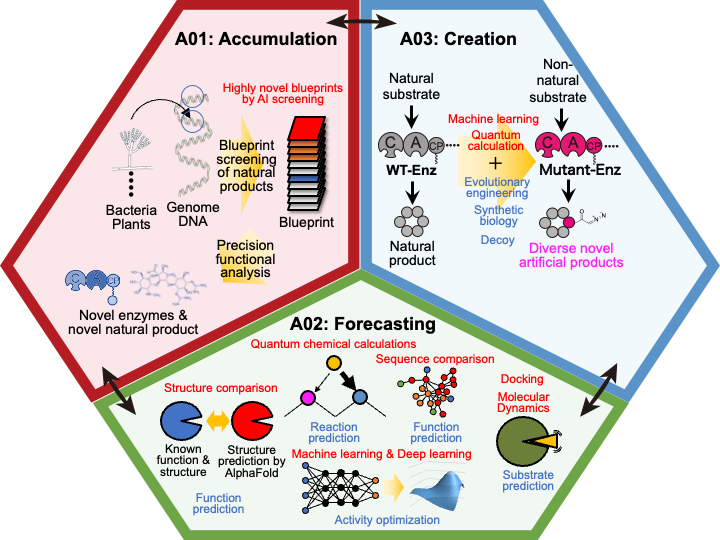
Research group A01 aims to identify novel biocatalysts that synthesize the basic framework of natural products by utilizing existing bioinformatics methods and AI (initially in progress) to be developed under research group A02.
Research group A02 aims to develop predictors for biological reactions by integrating structure prediction, machine learning, and quantum chemical computation, and to develop AI that can efficiently improve enzyme activity and extend substrate specificity.
Research group A03 aims to develop new molecular creation methods to extend the reaction of biocatalysts by synthetic biology and evolutionary engineering, AI, machine learning, molecular dynamics calculations, and quantum chemical calculations.

KUZUYAMA Tomohisa
University of Tokyo, Graduate School of Agricultural and Life Sciences, Professor
Expected Research Achievements
Natural products exhibit a variety of biological activities. Their blueprints, or biosynthetic genes, have become readily available in recent years. The "blueprints," which consist of thousands of ACGT nucleobase combinations, contain all the precursors (materials) and biosynthetic reactions (processes) to elaborate the natural products (final products) as a code. If so, could it not be possible to quickly predict the structure of a natural products by processing the biosynthetic gene sequence as "character information"? Can we create desired compounds by rationally modifying the biosynthetic genes or by designing them from scratch? In this Research Area, we aim to establish a methodology to predict "materials" and "final products" with high accuracy by deciphering the "process" described in the "blueprints," and to develop a predictor for such prediction. AlphaFold2, announced in the summer of 2021, can predict the steric structure of biosynthetic enzymes with overwhelmingly high accuracy based on sequence information alone, which is expected to cause a major paradigm shift in the prediction of biosynthetic enzyme functions. As a result, a paradigm shift is also about to occur in the prediction of biosynthetic enzyme functions. The advent of these ever-evolving predictors has greatly increased the realism of methods for predicting and pioneering the structures of unknown natural products.
In this Research Area, we aim to establish a new paradigm for biosynthetic science by making breakthroughs through diverse research spanning a wide range of fields, including experimental natural product chemistry, bioorganic chemistry, synthetic organic chemistry, synthetic biology, structural biology, theoretical computational chemistry, theoretical chemistry, computational biophysics, information science, artificial intelligence (AI), etc. More specifically, the project aims to establish a new paradigm in biosynthetic science; we will lead a fundamental change from the conventional concept, which has persisted for more than half a century in the field of natural product chemistry, that natural products are compounds to be "searched for," to the idea that natural products are compounds to be "created". In addition, this Research Area aims to nurture a younger generation of researchers who can freely use both experimental and theoretical approaches, rather than simply collaborating with each other, thereby contributing to the development of young researchers who will be indispensable in promoting digital transformation (DX) in various fields.


